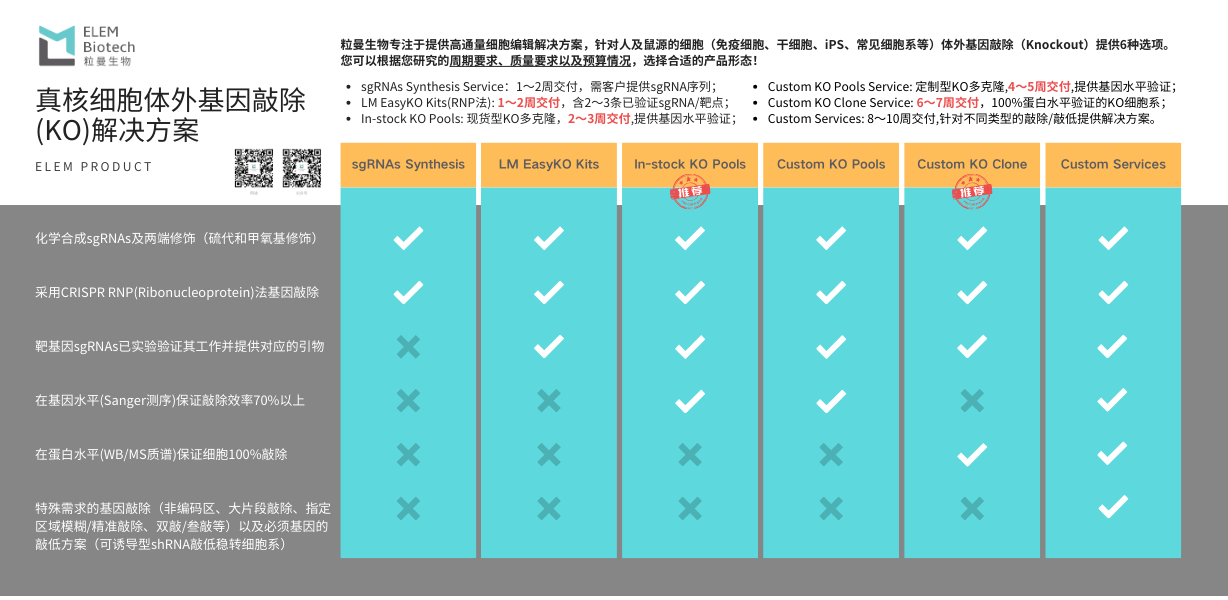
1. What is a KO Cell Pool?
A mixture of successfully edited and unsuccessfully edited cells obtained by transfecting a certain number of wild-type cell lines using gene editing methods (lentivirus, plasmid, RNP, etc.).
2. Why choose a KO Cell Pool?
(1) KO Pool preparation is faster and cheaper than cell lines, allowing for rapid validation of gene function;
(2) Genetic diversity: KO Cell Pools usually contain multiple different knockout cells. This diversity is closer to the genetic heterogeneity of cell populations in nature. This diversity can provide more comprehensive data and help researchers understand the impact of gene knockout on cell populations, rather than just the effects on a single cell line.
(3) Stability of experimental results: Due to the inclusion of multiple knockout cells, KO Cell Pools show higher stability and reproducibility in some experiments. In monoclonal cell lines, any unexpected variations or adaptive changes caused by gene knockout can lead to fluctuations in experimental results, while in cell pools, this effect can be balanced by diversity.
(4) Suitable for complex biological problems: For studying complex biological problems, such as multi-gene interactions and signaling pathways, using a KO Cell Pool may be more appropriate. Diversity ensures that different types of knockout cells can simulate complex biological processes and provide more physiologically relevant experimental conditions.
3. How to obtain a true KO Cell Line?
(1) Single clones obtained from KO polyclonal cells (KO Cell Pool) through serial dilution/flow cytometry/single-cell sorting techniques, and confirmed by Sanger sequencing to have a precise gene deletion or frameshift mutation, are truly KO monoclonal cell lines (KO Cell Line). KO Pools obtained using lentivirus and plasmid methods obtain monoclonal cells through drug screening, which only confirms plasmid integration into the genome and cannot confirm whether true gene editing has occurred. Further experiments are still needed to verify whether true gene editing has occurred.
(2) What is a homozygous KO cell line? All alleles of the wild-type cell undergo effective gene editing (non-3-fold frameshift mutations or fragment loss exceeding 21 amino acids), and no wild-type gene exists; this is a homozygous knockout cell line. Due to the probabilistic nature of gene editing, it is difficult to achieve completely identical editing sites at the allele level, but the absence of a wild-type gene represents successful homozygosity.
The CRISPR/Cas9 RNP gene editing method mixes multiple in vitro chemically synthesized single-guide RNAs (sgRNAs) corresponding to the target gene with Cas9 protein in vitro to form a ribonucleoprotein complex (RNP). Using electroporation/lipid transfection, the RNP is delivered into the cell in a process similar to "transient" transfection, without the need for the traditional lentivirus/plasmid resistance screening process, to obtain highly efficient and stably inherited cell pools. Since the RNP is degraded after completing genome editing within 72 hours in the cell and does not introduce exogenous sequences, it maintains the cell phenotype as much as possible, representing the method with the lowest off-target effect. It effectively avoids random integration of the viral genome, affecting subsequent functional experiments, and avoids the high off-target risk caused by continuous expression of Cas9 and sgRNA, thus maintaining cell genomic stability. CRISPR/Cas9 RNP gene knockout cell line construction is underway and will completely replace traditional methods. The figure below shows the basic workflow of CRISPR/Cas9 RNP gene editing.
Liman provides custom eukaryotic cell in vitro gene knockout services. For this service, the Liman team will regularly update the Q&A to better serve our customers. The Q&A [2409 version] is as follows:
1. What information do I need to provide when inquiring about gene knockout needs?
You only need to provide the cell name and target gene ID (it must be a unique ID, such as Gene symbol, Gene ID, Ensembl ID). We will query the gene essentiality (ES value) and the gene's expression in the target cell using the Liman database, providing you with relevant evaluation results (whether gene knockout or knockdown is recommended).
2. What does the ES value represent?
Based on Liman's unique gene editing database (not publicly available), it contains an assessment of the impact of each gene on cell growth in over 700 cell lines. The ES value is used to determine whether knocking out the target gene in a specific cell will affect cell growth. The higher the ES value, the greater the impact on cell growth after knockout, and the more difficult it will be to obtain a stable cell line.
3. Are there any requirements for the range of cell types used for gene knockout?
We can edit human/mouse immune cell lines/tumor cell lines/normal cell lines/primary cells/stem cells, etc., for in vitro functional studies; according to international ethical principles, we do not undertake cell editing for in vivo human experiments.
4. If I am going to do a gene knockout project, do I need to provide the relevant cells?
There are three ways for our project to obtain cells. First, customers provide them (they must be free of mycoplasma contamination; additional testing and culturing fees apply). Second, use Liman Biological Cell Bank's cells. Third, we have fixed and reputable cell suppliers who can procure cells for you.
5. What is the overall service cycle for gene knockout?
Taking cell A549 (doubling time ~22h) as an example, the cells are thawed, the cell status is adjusted to allow for electroporation, electroporation is arranged to obtain KO efficiency, and KO Pool is expanded and plated to obtain single clones.
Overall, our company obtains the KO Pool in about 2-4 weeks and the KO single clones (homozygotes) in about 8-10 weeks. The cycle of related projects will vary depending on the doubling time of the cells.
6. What is the difference between KO Pool (polyclone) and KO Cell line (monoclone)?
The KO Pool is a polyclone formed after knockout of wild-type cells. The genotypes of the gene knockout cells in the Pool are different, with fragment deletions or frameshift mutations, and may also contain a small number of wild-type cells. The knockout efficiency of the Pool can reach 100%, but the genotype is not unique. A monoclonal cell line has a completely consistent genotype with 100% knockout efficiency.
7. What is the principle of your gene knockout method?
We use CRISPR RNP (Cas9 and sgRNA complex) instead of traditional lentivirus and plasmid methods to improve editing efficiency while avoiding off-target effects caused by genome recombination. Using our leading sgRNA design, we provide editing efficiency analysis services after editing, freeing you from tedious virus packaging, plasmid preparation, Cas9 concentration testing, and other optimization experiments, allowing you to focus on biological discovery.
What are the principles of Cas9 target site screening?
The design of the particle man has three points: First, target the early exons of the coding functional protein; Second, target the shared exons of multiple isoforms of the target gene; Third, perform low-off-target analysis of the genome.
What exactly is RNP electroporation?
Specific sgRNAs are designed for the protein of interest, and the sgRNA and Cas9 complex (RNP) are transfected into cells using electroporation.
The sgRNA targets Cas9 to a specific site on the chromosome, causing frameshift or fragment loss in the corresponding gene.
When performing KO electroporation, are multiple sgRNAs mixed together or transfected individually?
According to different targets, we will design 2-3 highly specific sgRNAs with low off-target effects and mix them together for electroporation.
Will transfecting multiple sgRNAs together lead to higher off-target effects?
Studies have shown that the off-target efficiency of the RNP method is much lower than that of the viral and plasmid methods. First, the sgRNA design targets high specificity. Second, the RNP has a short half-life of only 72 hours and will degrade quickly, so the probability of off-target is low. This is also one of the advantages of the RNP method over lentivirus.
There are no fluorescent and resistance genes after RNP transfection, how to judge the transfection?
The RNP method is very efficient. After transient transfection (electroporation), editing begins and fluorescent/resistance screening is not needed. Stable KO polyclonal cell lines can be obtained after 3 days. Since the knockout efficiency of the pool is very high, generally reaching more than 70%, it is highly efficient to isolate monoclonal knockout homozygotes.
Electroporation was used for knockout. Was electroporation also used for the control group?
The control group does not need electroporation. Electroporation is only a transient perforation (tens of milliseconds) for the cells, and the cell membrane has fluidity and can recover quickly. Numerous studies have shown that electroporation has better cell tolerance compared to liposome transfection. In other words, for gene knockout using the RNP method, the control group cells are wild-type cells and no empty plasmid needs to be prepared for co-transfection.
What method was used to obtain the gene knockout results?
Using our company's optimized sgRNA design software, suitable target sites are screened, and multiple sgRNAs are designed for each target gene to increase the probability of multi-site cutting and improve the knockout efficiency; PCR primers matching the sgRNA sequence are automatically generated to amplify the genome editing site, and then the Sanger sequencing results are analyzed using the particle man's proprietary software to obtain the post-editing KO genotype analysis results.
Is the sequencing performed on the knocked-out segment, or on the entire gene?
Our designed primers only amplify the editing sites. One PCR product can contain all the sgRNA cutting and editing sites, thus determining the editing efficiency of the target. We currently do not provide whole-genome sequencing.
What is included in the delivery of your gene knockout project?
Our cell delivery standard is one tube of cryopreserved KO cells and related gene knockout reports (including gene knockout efficiency and cell culture methods) and related sequencing raw data. In addition, the particle man provides protein-level verification results, using WB verification or proteomics verification methods.
If you need more KO cells, we have corresponding charging standards, which can be consulted with our sales.
After obtaining the KO Pool, how are monoclonal strains screened without antibiotics?
For KO Pools with high KO efficiency and genetic stability, most of them can be directly used for phenotype screening and testing. If monoclonal cells are needed, they can be obtained through conventional monoclonal culture methods, such as limiting dilution, flow cytometry sorting, or single-cell microfluidic separation.
Are the delivered monoclonal cells homozygous?
According to our company's quality control standards, homozygotes are those where both alleles are knocked out (large fragment deletion or frameshift). The delivered monoclonal cells are homozygous if the genome does not contain wild-type sequences. The frameshift or fragment loss in the alleles of the delivered homozygous cells cannot be guaranteed to be completely consistent, but it can be guaranteed that there is no wild-type protein expression.
If the cells I received in one tube of cryopreserved cells are not successfully cultured after resuscitation, will you provide them again?
Each project cell we deliver has a backup. If you fail to resuscitate the cells, we can provide you with one tube of cryopreserved cells free of charge within one month of your receiving the cells, and we can also provide you with technical support for cell culture.
After knockout, there are still bands in the WB experiment results. Why does this happen?
If you are working with a KO Pool: From a genotype analysis, it contains 15% wild-type sequences, which may be the reason for the weak bands. The KO Pool can be screened for monoclonal cells to obtain stably inherited monoclonal cell lines. Also note that some genes have similar transcripts or highly homologous proteins. The specificity of the target gene and the verification antibody used needs to be considered comprehensively.
For more details, please refer to:
Problem Interpretation I Why WB Experiments Are Not Completely Suitable for Evaluating the Knockout Efficiency of KO Cells (1)
Problem Interpretation I Why WB Experiments Are Not Completely Suitable for Evaluating the Knockout Efficiency of KO Cells (2)
Problem Interpretation I Why Proteomics Verification Is the Gold Standard for KO Cell Knockout Verification? [Collection]
Service Guide I Particle Man High-Throughput Gene Knockout Custom Service (RNP Method) Q&A [2409 Version]