Product Launch | "Ubiquitination Enrichment Antibodies" — A Product Line in the Protein Degradation Drug Field [Save This!]
2025-04-18
【Liman's "Protein Degradation Drug Portfolio" Officially Launched!】
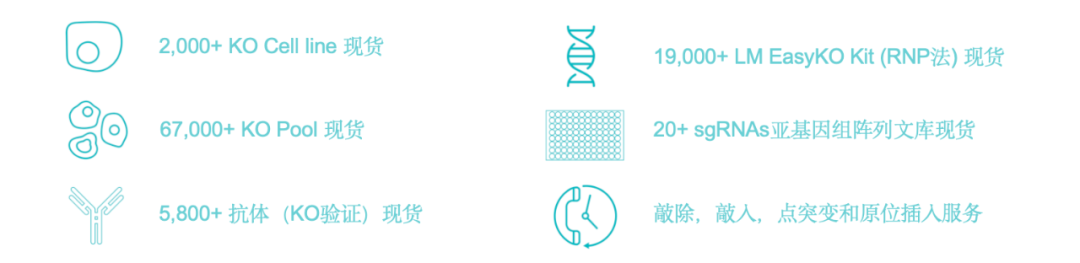
The team at Geman Bio has been deeply engaged in the development of protein degradation drugs, offering comprehensive services that span from target discovery and validation, to lead compound identification and optimization, as well as drug mechanism-of-action (MOA) studies and PK/PD analyses. Geman provides high-throughput gene-editing products—such as KO/KI cell lines—as well as complementary mass spectrometry-based reagents and personalized services, helping to streamline and accelerate the research process. Through our diverse product portfolio in this field, we deliver end-to-end, tailored solutions to both pharmaceutical clients and researchers in basic science.
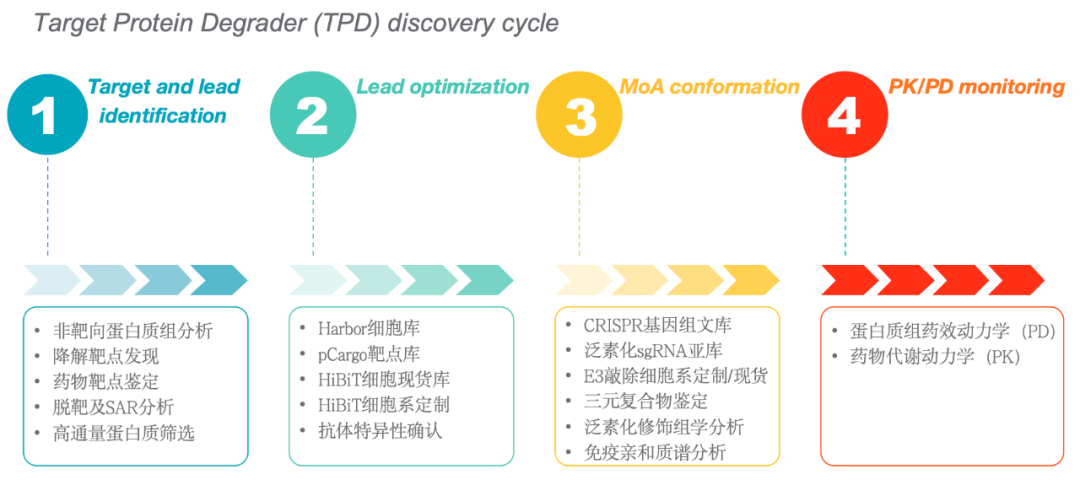
Figure 1: Geman's Product and Service Portfolio in the TPD Field
1. Protein Ubiquitination and Identification of Ubiquitination Patterns to Determine the Mechanism of Action (MOA) of Proteolysis-Targeting Chimeras
Ubiquitination of proteins is a post-translational modification process mediated by E3 ubiquitin ligases, which targets specific proteins for degradation via the proteasome system (UPS), thereby maintaining cellular homeostasis. In recent years, this critical biological process has been harnessed through small-molecule compounds—such as PROTACs or molecular glues—to enhance the interaction between E3 ubiquitin ligases and disease-associated target proteins (POIs), promoting the ubiquitination and selective degradation of these disease-driving proteins, ultimately achieving therapeutic benefits. As an increasing number of protein-degrading drug candidates advance into preclinical research and clinical trials, ubiquitination-based proteomics offers invaluable insights by specifically identifying and quantifying the extent and dynamic changes in POI ubiquitination. This not only deepens our understanding of how these compounds exert their mechanisms of action and therapeutic efficacy but also provides direct data support for optimizing structure-activity relationships (SAR). Moreover, ubiquitination proteomics can help pinpoint potential off-target effects of targeted protein degraders by analyzing ubiquitination patterns in cellular or tissue samples, thereby reducing the risk of adverse clinical outcomes caused by off-target interactions.
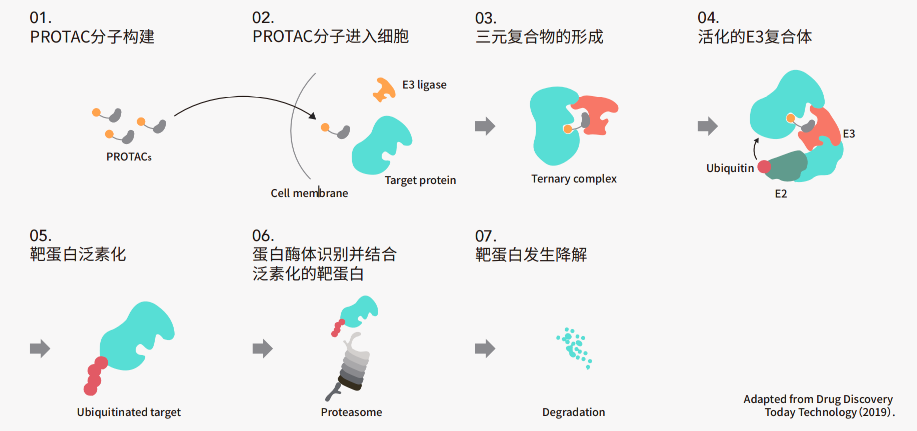
Figure 2: Mechanism of PROTAC Degradants
2. K-diGly Antibody, independently developed by Leman
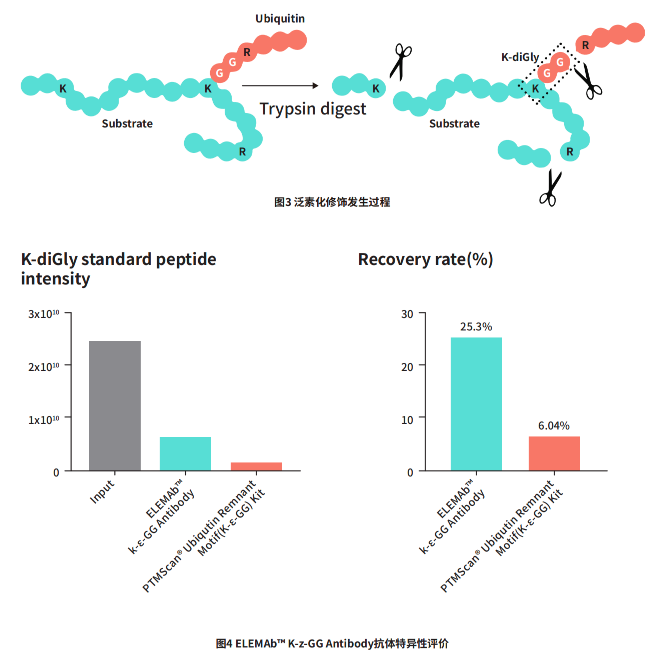
Ubiquitination refers to the process in which the ε-amino group of a lysine (K) residue in the substrate protein forms an amide bond with the carboxyl group of the terminal glycine in the diGly moiety of a ubiquitin molecule. After trypsin cleaves arginine (R) and lysine (K), a characteristic K-GG sequence is generated. Given that ubiquitinated peptides typically exhibit low abundance and a wide dynamic range, it is essential to enrich these modified peptide segments prior to detection for subsequent mass spectrometry analysis. To this end, we first performed animal immunization using ubiquitinated peptides and successfully obtained the ELEMAb™ K-ε-GG Antibody, specifically tailored for detecting ubiquitination modifications. We then utilized the SpikeMix™ PTM-Kit 47-Lys (GG) to evaluate the enrichment efficiency of standardized ubiquitinated peptide segments.
3. High-Repeatability Ubiquitination Modification Omics
Over 96.54% of ubiquitinated proteins and 93.51% of ubiquitinated peptides were repeatedly identified, demonstrating the excellent reproducibility of the ELEMAb™ K-ε-GG Antibody in ubiquitin modification analysis. These results highlight the antibody's reliability and robustness in detecting and enriching ubiquitinated modifications, providing a solid foundation for future research.
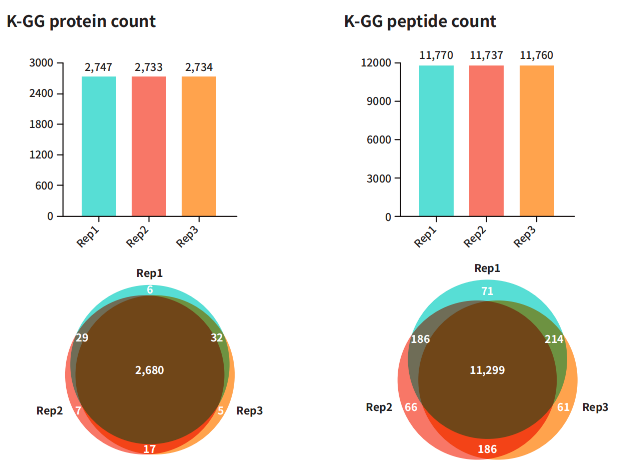
Figure 5: Reproducibility Evaluation of ELEMAb™ K-ε-GG Antibody Identification
4. Ubiquitination Proteomics Confirms the Mechanism of Action of Degradants
Lenalidomide is a drug that demonstrates significant clinical efficacy against multiple myeloma and other B-cell malignancies. To gain deeper insights into the mechanism of action of lenalidomide, researchers employed ubiquitination proteomics and discovered that lenalidomide leads to increased levels of ubiquitination for both IKZF1 and IKZF3. Furthermore, comprehensive proteomic analysis revealed that lenalidomide reduces the abundance of these two proteins. As key transcription factors in multiple myeloma, this finding sheds light on how lenalidomide exerts its anti-tumor effects by modulating the ubiquitination and subsequent degradation of IKZF1 and IKZF3.
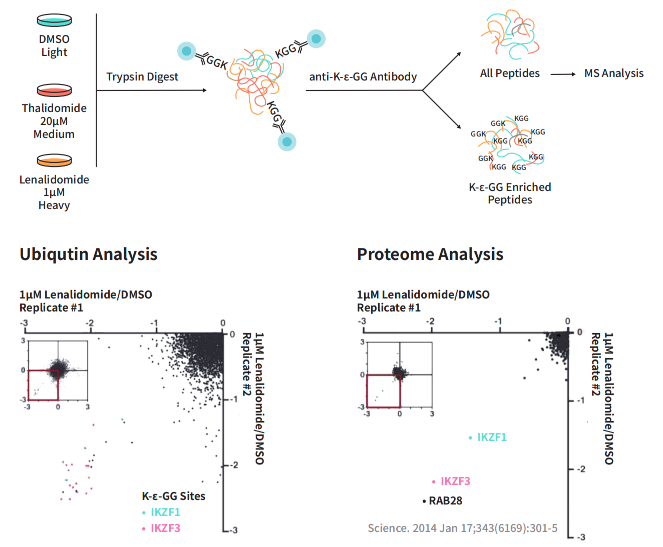
Figure 6: Ubiquitination Omics Reveals the Mechanism of Action of Lenalidomide
References
1. Targeted protein degradation mechanisms. Drug Discov Today Technol. 2019 Apr;31:53-60.
2. Lenalidomide induces selective degradation of IKZF1 and IKZF3 in multiple myeloma cells. Science. 2014 Jan 17;343(6168):301-5.
2025 /
04-18
Classification:
Company News
Related Information