Technical Topic | Cracking the Code of Intracellular Protein Interactions: The Value and Selection Strategies of Proximity Labeling Technologies in Biological Research【Collection】
2025-04-14
Background
In the post-genomic era, understanding the complex and dynamic interaction networks between proteins within cells has become key to revealing the mysteries of life. Proximity Labeling Technology, as an emerging method for capturing protein interactions, is becoming a powerful tool in life science research due to its high spatiotemporal resolution and suitability for live cells/organisms.
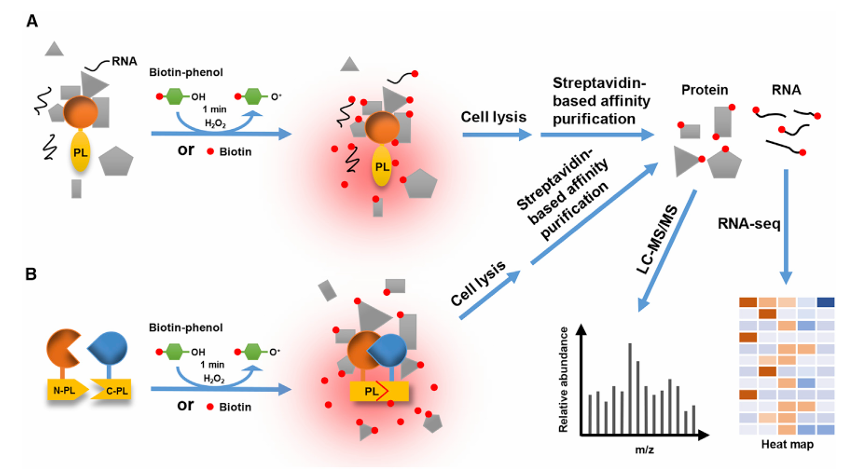
Proximity Labeling Flowchart [1]
This article focuses on the advantages and disadvantages of different construction strategies of proximity labeling technology, helping you accurately pinpoint your research design.
I. Introduction to Proximity Labeling Technology
Proximity labeling is a protein interaction detection method based on engineered enzymes. Its core principle is to fuse a catalytically active labeling enzyme (such as TurboID, APEX2, miniTurbo, AirID, etc.) with the target protein for expression. Under specific conditions, it catalyzes the covalent attachment of capturable labels (such as biotin) to nearby proteins (within a 5-10 nanometer range). Through streptavidin enrichment and mass spectrometry identification, the microenvironment of the target protein can be analyzed.
Supplementary Reading: Literature Interpretation | AirID: A New Chapter in Protein Interaction Research [Collection]
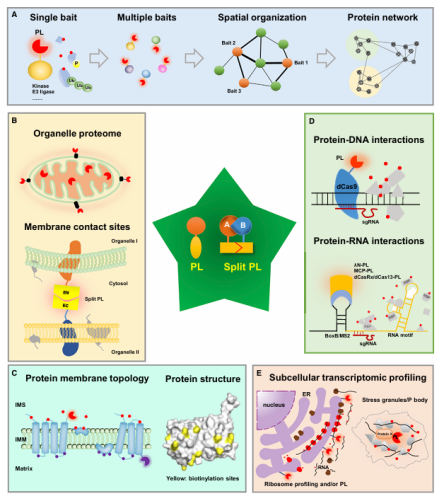
II. Research Value and Typical Applications of Proximity Labeling
1) Constructing protein interaction maps (Interactome Mapping): Revealing the proximity network of specific proteins or complexes, supplementing the blind spots of traditional co-immunoprecipitation.
2) Subcellular localization microenvironment analysis: Locating the labeling enzyme in organelles (mitochondria, lysosomes, etc.) or membrane structures to analyze region-specific protein composition.
3) Protein transport and signal dynamic tracking: Studying changes in the components of protein complexes under stimulation or drug action.
4) In vivo animal interaction research: Utilizing the low toxicity of TurboID/miniTurbo to study tissue-specific interactions in mice or zebrafish.
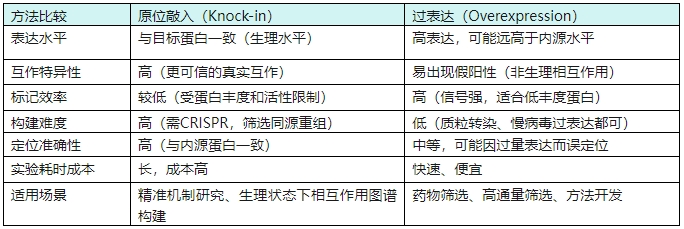
III. Cell Line Construction Strategy Selection: In-situ Knock-in vs. Overexpression Fusion
There are two main methods for constructing proximity labeling systems in cell lines:
3.1 Overexpression Fusion (OE)
3.1.1 Principle
The fusion protein (e.g., TurboID-target protein) is introduced into cells through an exogenous expression system (such as plasmids, electroporation, viruses, etc.), allowing it to exist at a high expression level, usually with random insertion.
3.1.2 Construction Process
1) Clone the target protein + proximity labeling enzyme (such as TurboID, APEX2) fusion construct into an expression vector.
2) Introduce into cells using plasmids, lentiviruses, adeno-associated viruses (AAV), etc.
3) After expression confirmation, add substrate (such as biotin or biotin-phenol) for labeling experiments.
3.2 In-situ Insertion (Endogenous Knock-in)
3.2.1 Principle
Through methods such as CRISPR-Cas9, the TurboID/APEX proximity labeling enzyme gene is inserted in a fused form into the endogenous gene locus (N-terminus/C-terminus of the target gene) to achieve proximity labeling regulated by endogenous expression.
3.2.2 Construction Process
1) Design sgRNA targeting the stop codon of the target gene, i.e., C-terminal insertion (or after the start codon - N-terminal insertion).
2) Prepare donor DNA containing TurboID or other tags and homologous arms. 3) Electroporate or use viral transduction to introduce into cells and screen for successful knock-in clones. 4) Verify the expression and functional integrity of the fusion protein.
Comparison of Proximity Labeling Construction Methods
IV. Reference Cases
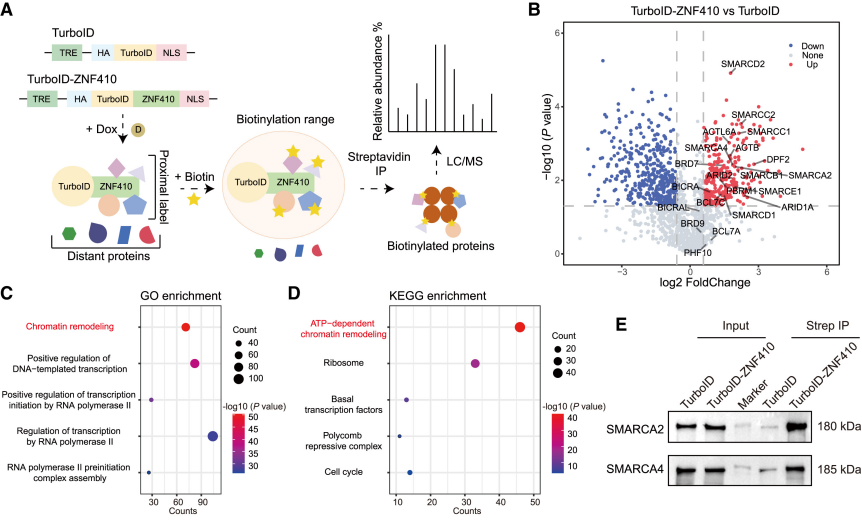
4.1 Interaction Protein Screening (Overexpression)
Researchers overexpressed TurboID fused with ZNF410 Overexpression Fusion [2] to achieve biotinylation of proteins around ZNF410, confirming that the chromatin remodeling complex SWI/SNF is selectively recruited by the N-terminus of ZNF410. The two work synergistically to evict nucleosomes, ensuring efficient binding of ZNF410 to HCTs in the enhancer region, thereby maintaining the open chromatin state of the CHD4 enhancer.
4.2 Studying Disease Mechanisms (Overexpression)
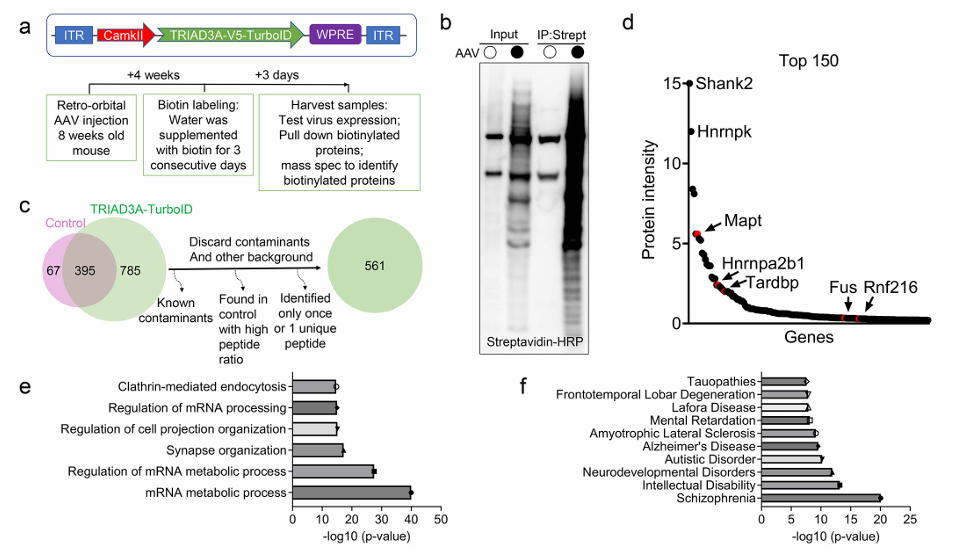
Researchers have discovered a novel mechanism regulating tau pathological aggregation [3] : TRIAD3A promotes tau amyloidogenesis through nested liquid-liquid phase separation . The experiment used TurboID Overexpression Fusion TRIAD3A and labeled its neighboring proteins in live cells, identifying multiple proteins associated with neurodegenerative diseases, including tau. This study not only highlights the importance of the hierarchical organization of phase separation structures in pathological protein aggregation; it also provides new insights into the connection between the ubiquitin system, phase separation, and autophagic clearance, providing potential targets for interventions in diseases such as Alzheimer's disease.
4.3 Constructing Interaction Maps (Overexpression & In-situ Knock-in)
Researchers comprehensively compared the performance differences between TurboID in-situ knock-in (KI) and overexpression (OE) in proximity labeling experiments paying particular attention to signal-to-noise ratio, the number of interacting proteins, and specificity and other indicators [4] 。
1) Comparison of Biotin labeling efficiency revealed that the biotin signal was very strong in OE samples, but also accompanied by more background (non-specific labeling); In KI samples, the biotin signal was clear, with less background, and the distribution was closer to the target area.
2) Analysis of the interacting proteins identified by the two methods showed that the KI group showed a series of highly significant true interacting proteins with less background protein; while the OE group detected more proteins, but the high background masked the true interactions. 3) Functional enrichment analysis of the interacting proteins identified in the two groups proved that the biological processes enriched in the KI group were highly consistent with the function of the target protein (such as intranuclear processes, chromatin regulation, etc.), while the OE group showed a large number of irrelevant pathways (such as extracellular structural proteins, etc.).
4) Finally, assessing the overall quality of the MS data revealed that the KI method had higher sensitivity.
In summary, the researchers believe Endogenously expressed TurboID KI can provide a more specific, accurate, and physiologically relevant protein interaction map, which is a better strategy than the traditional OE method. 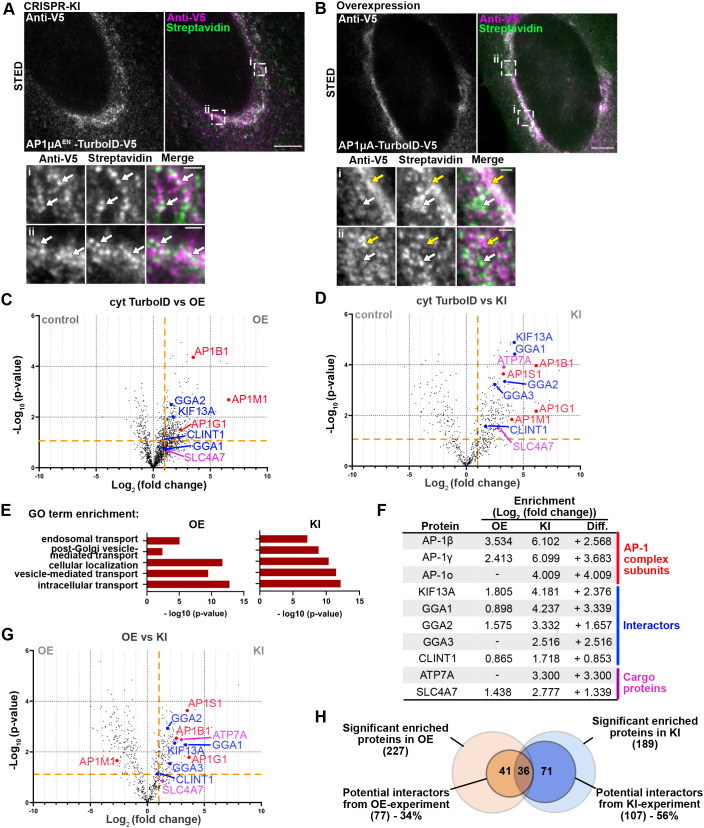
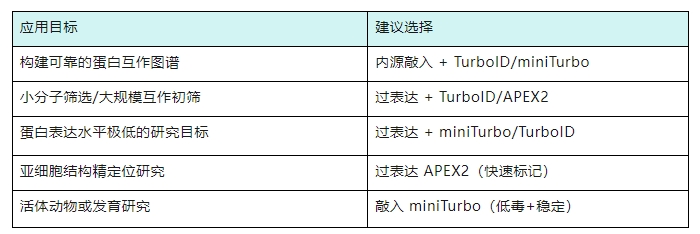
In summary, we recommend different strategies according to different application scenarios for your reference:
Liman provides a full range of services for proximity labeling (including overexpression/in situ labeling strategies, strep affinity purification, and mass spectrometry detection). Please contact us if you have any needs!
Liman's products and services related to "protein proximity labeling":
1. Gene in situ insertion/overexpression stable cell line construction : In situ insertion of birID, TurboID, etc., tags at the N-terminus/C-terminus of genes for proximity labeling detection, which can detect protein-protein interactions (weak interactions, transient interactions); in situ insertion of Strep-tag, etc., for AP-MS detection, which can detect strong protein-protein interactions at basal levels; random insertion or safe harbor site-specific insertion of "tag-target protein" to construct overexpression cell lines for PL research;
2. Liman strep II purification filler : For Strep-tag proteins and biotin-labeled proteins (e.g., obtained after proximity labeling) for ultra-high affinity enrichment (100 times higher affinity than antibody enrichment);
3. AP-MS/IP-MS detection : Mass spectrometry detection of cell samples with proximity labeling or affinity labeling (Strep-tag) to identify interacting proteins;
4. Human/mouse gene knockout arrayed library screening : For 19+ metabolic pathways (including 300+ E3 ubiquitin ligase KO array library, 1000+ ubiquitin-related enzyme KO array library), higher accuracy, simplified data interpretation, strong experimental reproducibility, and broader research needs.
V. References
[1] Yang X, et al.Proximity labeling: an emerging tool for probing in planta molecular interactions. Plant Commun. 2020 Dec 15;2(2):100137.
[2] Xu S, et al.SWI/SNF complex-mediated ZNF410 cooperative binding maintains chromatin accessibility and enhancer activity. Cell Rep. 2025 Mar 28;44(4):115476.[3] Zhou J, et al.The autophagy adaptor TRIAD3A promotes tau fibrillation by nested phase separation. Nat Cell Biol. 2024 Aug;26(8):1274-1286.[4] Stockhammer A, et al. When less is more - a fast TurboID knock-in approach for high-sensitivity endogenous interactome mapping.J Cell Sci.2024 Aug 15;137(16):jcs261952.
2025 /
04-14
Classification:
Industry News
Related Information

