Technical Topic | Protein Proximity Marker (PL): New Standard for Protein Interaction Research [Collection]]
2024-09-24
The function of proteins in organisms often depends on their interactions with other molecules, including-protein, protein-nucleic acid, protein-small molecule, etc. Traditional protein interaction detection methods, such as co-immunoprecipitation (Co-IP), GST Pull-Down, etc., provide effective tools for studying these interactions, but they usually have limitations such as low spatial resolution, harsh conditions, and damage to the integrity of organelles. Protein proximity labeling (proximity labeling, PL) technology has gradually become an efficient, specific and high spatial and temporal resolution tool for protein interaction research in recent years.
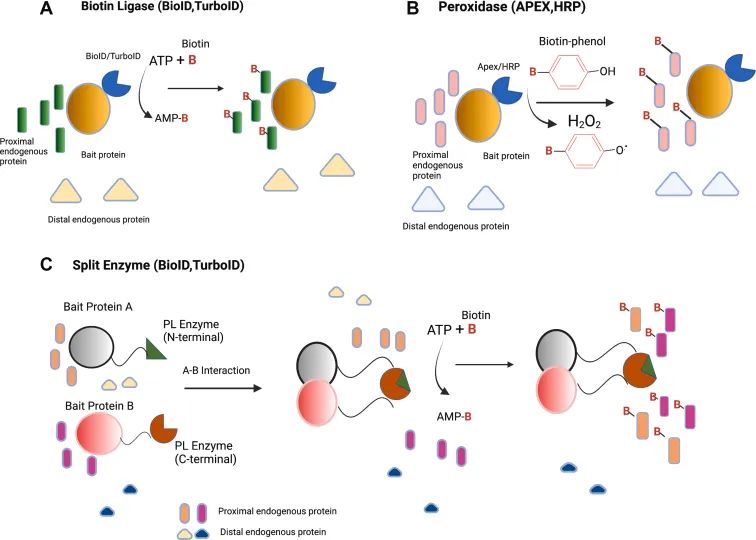
Figure1 Schematic diagram of labeling principle of commonly used PL enzymes (Mathew et al., 2022)
The Principle of Proximity Labeling of 1. Protein
The basic principle of protein proximity labeling technology is to fuse and express a certain enzyme (such as peroxidase or biotinylated enzyme) with the target protein. When the target protein is close to other proteins, the enzyme will trigger a chemical reaction to connect the active labeling molecule (such as biotin or fluorescent molecule) to the nearby protein. These tagged proteins can then be captured and identified by methods such as affinity purification, thereby revealing the interaction network of the target protein.
2. commonly used enzyme systems
1.BioID (biotinylation labeling method)
The BioID technology uses a mutant BirA * of the biotin protein ligase BirA, which is capable of randomly attaching biotin to proteins close to it (in the range of about 10nm) at low concentrations of biotin. These biotinylated proteins can then be enriched by streptavidin purification techniques and identified by mass spectrometry analysis.
2. APEX (peroxidase labeling method)
APEX (Enhanced Non-Natural Peroxidase) is another widely used proximity labeling system. The APEX enzyme is able to catalyze the oxidation of biotinophenol derivatives by hydrogen peroxide to reactive free radicals, which rapidly react with neighboring proteins to form covalent bonds. The labeling range of APEX is within 1-2nm, the labeling efficiency is high, and it can be used in living cells and fixed cells.
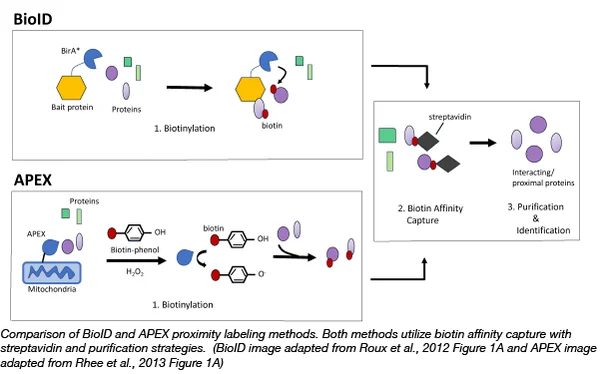
Figure2 BioID and APEX Comparison
3. TurboID和miniTurbo
TurboID is an upgraded version of BioID, with a shorter reaction time (labeling within 10 minutes), suitable for rapid labeling of living cells. The MiniTurbo is a miniaturized version of the TurboID, which also has efficient marking performance in a small space requirement and is suitable for specific experimental designs.
Application of 3. Protein Proximity Markers
1. Protein interaction network analysis
By proximity labeling technology, the target protein can be captured in situ in living cells."neighbors", thereby mapping their interaction networks. This method not only identifies stable complexes, but also detects transient or weak interactions.
researchers by combiningProximity labeling, quantitative mass spectrometry of APEX2 analyzed the kinetics of the proximity proteome of EGFR in the time dimension. A protein network involved in the signal transduction process is interpreted using this approach. Opening the way for the discovery of new regulatory mechanisms for signal transduction and intracellular trafficking of receptor tyrosine kinases.
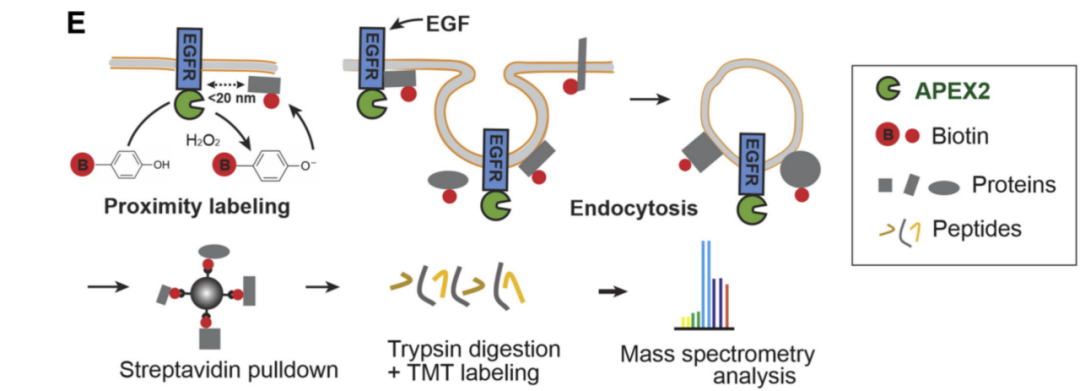
2. Subcellular localization and dynamic changes
Protein proximity labeling can be performed in specific organelles or cellular compartments, helping to study the distribution of proteins in specific subcellular structures. especiallyThe high spatial and temporal resolution of APEX can be used to monitor the dynamic changes of proteins in cells in real time.
Researchers useAPEX2 is adjacent to the labeling of the copper chaperone protein Atox1, which is then used as a "bait" using the nuclear localization and copper transport functions of this protein, thus providing a systematic view of the copper protein interaction network with spatial and temporal resolution in living cells. Through the quantitative proteomic analysis of dimethyl labeling, CRIP2 can be used as a potential copper binding protein and Atox1 interaction, providing a new mechanism of action and drug screening targets for the treatment of copper disorders in tumors.
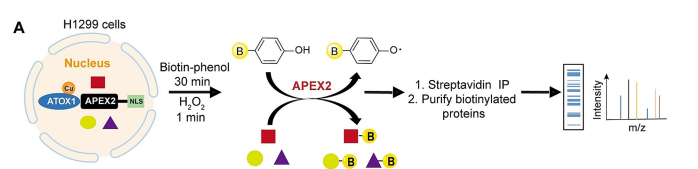
3. Identification of specific protein complexes
In some biological processes, the function of proteins often depends on the complexes they form. Proximity labeling technology can capture these complexes and identify them by mass spectrometry, providing clues for understanding complex signaling pathways or regulatory mechanisms.
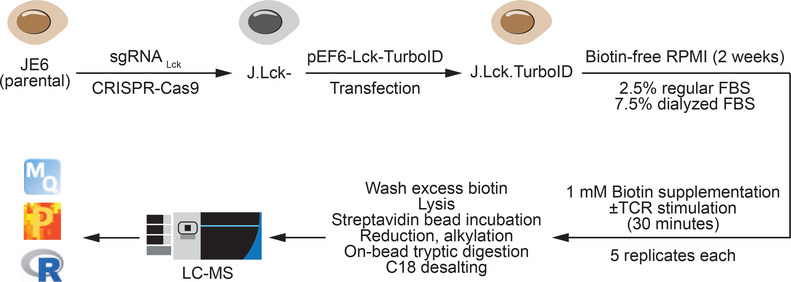
In the following study,TurboID was used to study the Lck kinase complex in human T cells, where Lck plays a crucial role in T cell receptor signaling. The researchers TurboID captured the proteins that interact with Lck when T cells activate, revealing a key component of the signaling complex in the immune response.
4. Drug Target Discovery
ubiquitin-The proteasome system (UPS) is one of the major intracellular protein degradation systems, and the abnormal regulation of ubiquitin ligase mutations is associated with many diseases. The identification of E3 ubiquitin ligase substrates has been challenging due to its low affinity, short interaction, and easy target degradation. Especially in the field of protein degradation drugs (TPD) research, the identification of E3 ubiquitin ligase interacting proteins has become an area of increasing concern.

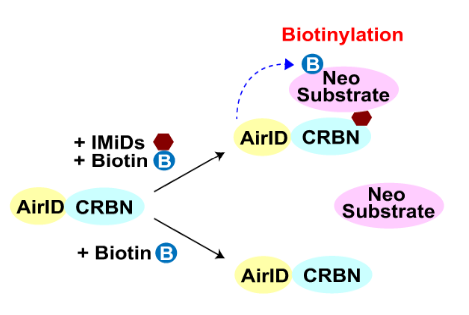
Comparison of 4. Common Protein Interaction Techniques

Co-IP are suitable for validating known stable protein complexes, but are less sensitive to transient and weak interactions.
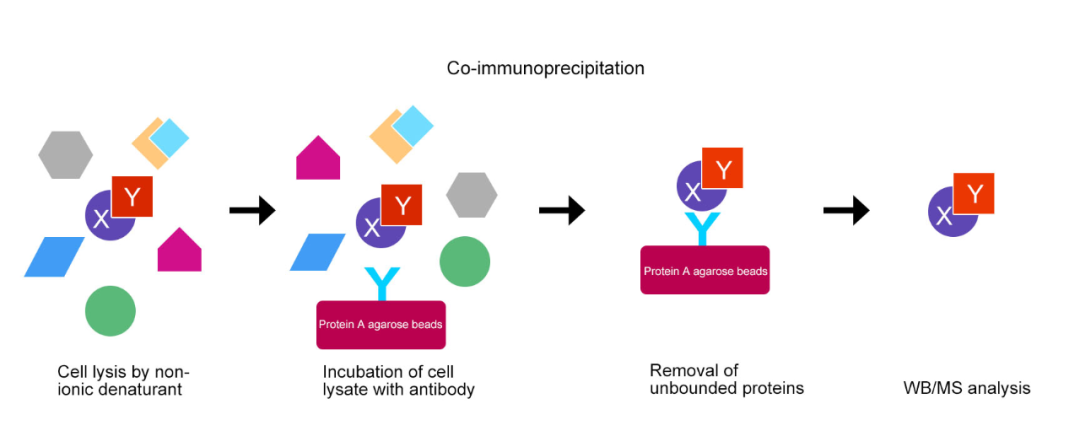
Figure3 Co-IP basic experimental process
GST pull-down is suitable for analyzing protein-protein interactions in vitro, especially for detecting known direct interactions, but not for studying complex, dynamic interactions.
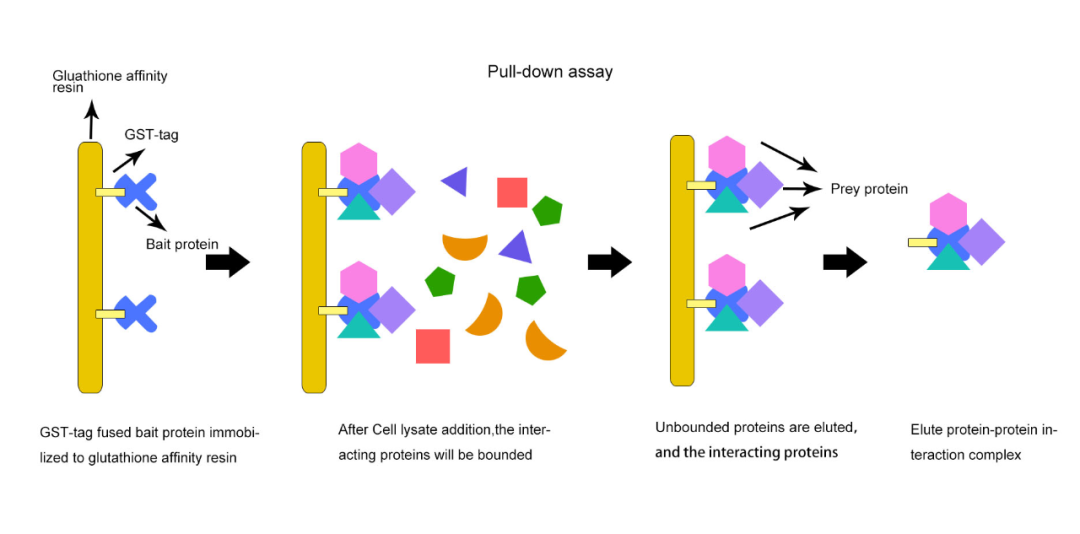
Figure4 GST pull-down basic experimental process
PL(proximity labeling) is carried out in vivo and is suitable for capturing dynamic, transient protein interactions, especially those of subcellular compartments or membrane proteins.
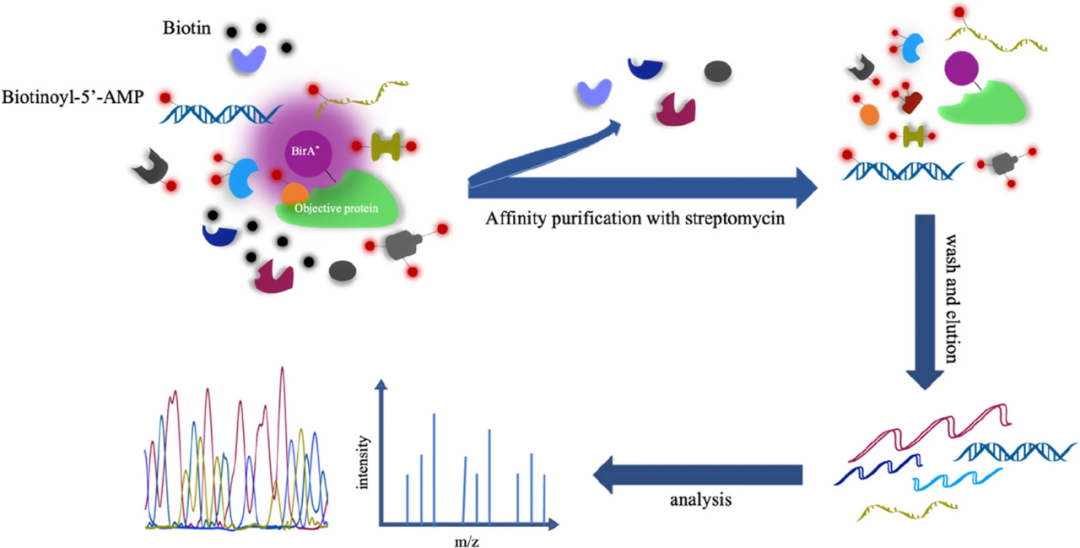
Figure5 PL(proximity labeling) basic experimental process
6. References
[1] Mathew B, Bathla S, Williams KR, Nairn AC. Deciphering Spatial Protein-Protein Interactions in Brain Using Proximity Labeling. Mol Cell Proteomics. 2022 Nov;21(11):100422. doi: 10.1016/j.mcpro.2022.100422.
[2] Perez Verdaguer M, et al.Time-resolved proximity labeling of protein networks associated with ligand-activated EGFR. Cell Rep. 2022 Jun 14;39(11):110950.
[3] Chen L, et al.APEX2-based Proximity Labeling of Atox1 Identifies CRIP2 as a Nuclear Copper-binding Protein that Regulates Autophagy Activation. Angew Chem Int Ed Engl. 2021 Nov 22;60(48):25346-25355.
[4] Chua XY,et al.Quantitative Interactomics of Lck-TurboID in Living Human T Cells Unveils T Cell Receptor Stimulation-Induced Proximal Lck Interactors. J Proteome Res. 2021 Jan 1;20(1):715-726.
2024 /
09-24
Classification:
Industry News
Related Information

